DATABASES

Network pharmacology is a new discipline that has emerged which attempts to understand drug action and interactions with multiple targets. It uses computational power to systematically catalog the molecular interactions of a drug molecule in a living cell. Network pharmacology appeared as an important tool in understanding the underlying complex relationships between botanical formula and the whole body. It also attempts at discovering new drug leads and targets and repurposing existing drug molecules for different therapeutic conditions by allowing unbiased investigation of potential target space. However, these efforts require some guidance for selecting the right type of targets and new scaffolds of drug molecules. Traditional knowledge can play a vital role in this process of formulation discovery and repurposing existing drugs. By combining advances in systems biology and network pharmacology, it might be possible to rationally design the next generation of promiscuous drugs with network pharmacology. Network pharmacology analysis not only open up new therapeutic options but also aids to improve the safety and efficacy of existing medications.
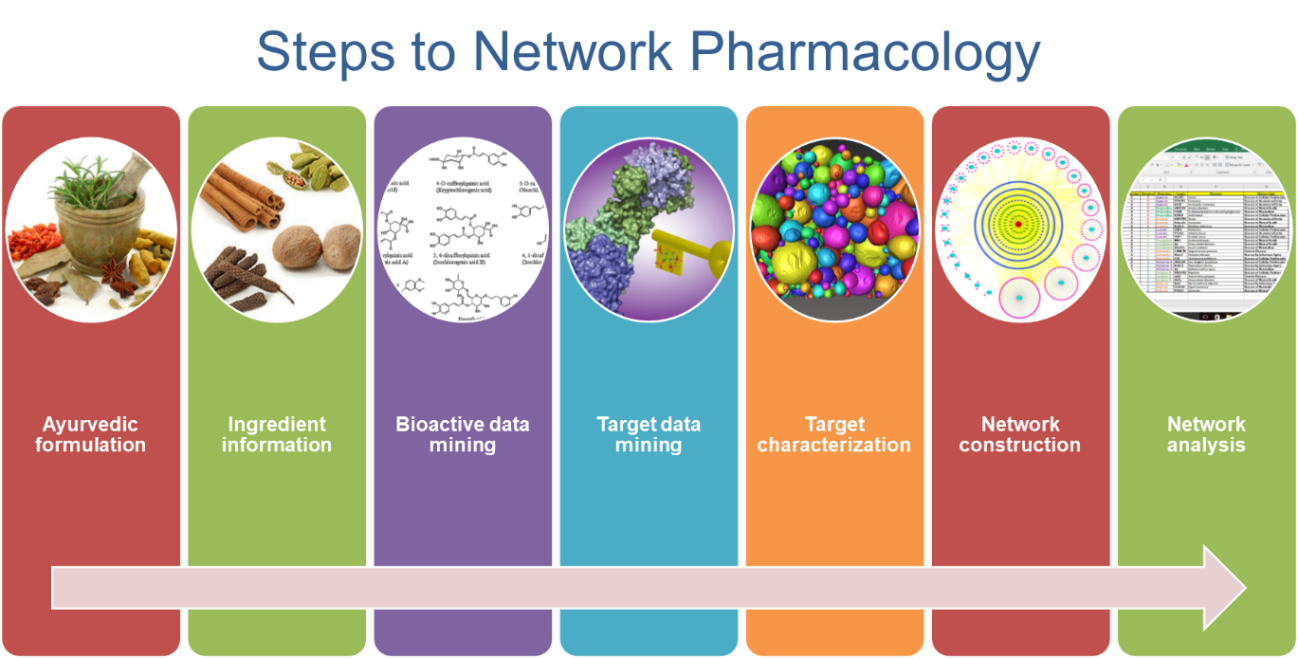
The diversifying activities of network pharmacology involves:
The steps of network pharmacology depend on objectives. However, the superficial and common steps involve-
What are HDIs?
A modification in the effect of one drug by another exogenous agent (modern drug, plant or food is termed as drug interaction. The risk for drug interactions increases with the number of products consumed: for 2 products, the risk is 6%; for 5 products, 50%; and for 8 or more products, 100% [1]. Hence in addition to evaluating safety and efficacy of herbal drugs, one needs to pay attention to their interactions with other concurrently administered drugs or food. AYUSH offers a wide range of Pharmacological-Non-Pharmacological remedies to manage the number of disease conditions. Ayurveda, Siddha, Sowa Rigpa and Unani, (ASU) drugs [2] [3] or herbal drugs mainly come under pharmacological interventions. ASU drugs hold significant capacity to alter the drug metabolizing enzymes (DMEs), and drug transporters (DTs). This poses a risk of drug-drug or herb-drug interactions (HDI) [4] [5]. The HDIs can be beneficial, harmful or even fatal. Therefore, it is a timely requirement to develop a methodology for screening of HDI to ensure the effective, safe, and rapid drug discovery and development pipeline. Many examples of adverse effects have been reported by Hu et al 2005 [6] and Borse et al. 2019 [4]. The clinically significant effects are usually associated with a doubling or more of plasma concentration of the modern drug [6]. However, if they are administered with agents with narrow therapeutic index (such as protease inhibitors, cytokines etc.) then even small changes may be clinically important.
In most cases, the drug interactions result because herbal drugs affect the biological processes that regulate metabolism and elimination of modern drugs, leading to changes in their pharmacokinetic parameters (like bioavailability, volume of distribution, protein binding, clearance and AUC). The herbal drugs may be substrates of the same metabolic and transport proteins, including cytochrome P450enzymes (CYP), glucuronosyltransferases (UGTs), and the efflux drug transporter P-glycoprotein (Pgp). Hence, studying HDIs is of paramount importance for analyzing efficacy and safety profiles of ASU drugs.
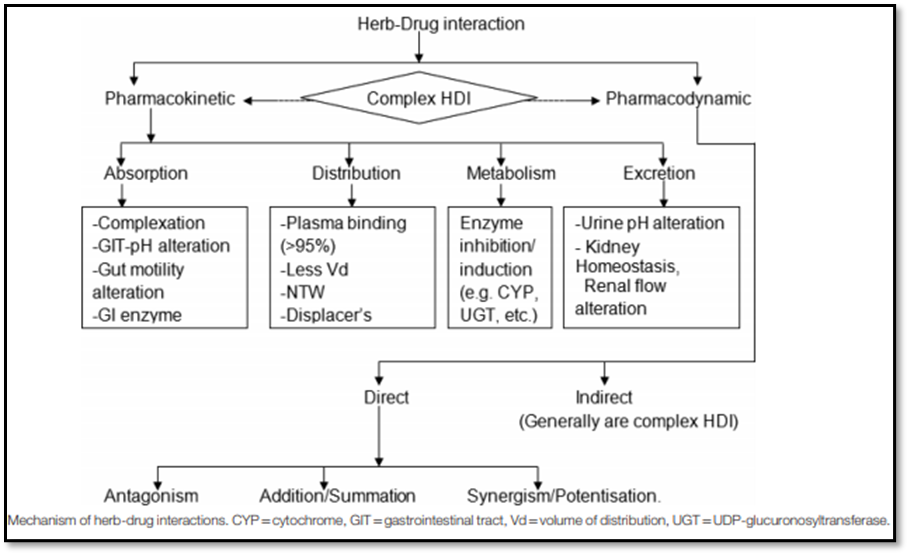
Mechanisms of HDIs?
HDIs are mediated by pharmacodynamic and/or pharmacokinetic mechanisms. Pharmacokinetic interactions are much more difficult to anticipate than pharmacodynamic interactions [7] [8]. Most commonly reported HDIs are pharmacokinetic interactions, especially those resulting from the functional modulation of DMEs, mainly cytochromes (CYPs); drug transporters such as P-gp; and protein binding. Pharmacodynamic interaction involves antagonism, addition/summation, synergism, and even sometimes modulation of drug targets [8] [9] [10]. The following figure describes the mechanisms of HDIs (Adapted from Borse et al. 2019).
How to study HDIs?
Different in-silico, in vitro, and in-vivo techniques for studying pharmacokinetics, toxicity and HDIs are as follows:
For developing any drug profile studying drug like properties, pharmacokinetics and toxicity and mode of action of drug substance is of great importance [11]. Analyzing these parameters at early stages is gaining attention nowadays to screen potential molecules or to decide appropriate strategies for the drug development process [12]. Conventional methods have cumbersome experimental procedures, are time consuming and expensive [13]. The evolution of computational approaches to optimize pharmacokinetic, toxicity and pharmacodynamics properties may enable the progression of discovery leads effectively and swiftly to drug candidates [14]. Few of the majorly used In-silico tools for pharmacokinetic and pharmacodynamics HDIs are as follows-
ADMET softwares:
There are several open-source web-based platforms to predict ADME. Each one of them works on different criteria, different algorithms and it uses different models for prediction, which makes them differ with respect to their effectiveness and accuracy. Some tools specifically predict only one property e.g., P-gp rules, SMARTcyp. Few of the tools are given in the following table-
| Tool | Availability | Batch computation | Database | Prediction(Total endpoints prediction) | Modelling Algorithm Method used | Drug-likeliness Rules | Systematic Evaluation |
|---|---|---|---|---|---|---|---|
| ADMETlab | free | Yes | Yes | Druglikeness, ADMET and Systematic Evaluation Number: 31 Contents: B, A, D, M, E, T | RF, SVM, RP and PLS for regression model SVM, NB and DT for classification model | Yes/Yes | Yes |
| ADMET-SAR | free | No | Yes | ADMET Number: 27 Contents: B, A, D, M, E, T | SVM methods: 22 classification models and 5 regression models Fingerprints: MACCS, Morgan and AtomParis | No/No | Yes |
| SWISS-ADME | Free | Yes | No | ADME Number: 19 Contents: B, A, D, M | SVM Bayesian technique BOILED-Egg model | No/No | Yes |
| pkCSM | Free | Yes | No | ADMET, Pharmacokinetics, Toxicity Number: 30 Contents: B, A, D, M, E, T | Cutoff Scanning algorithm | No/No | Yes |
| PreADMET | Free or com‑ mercial | No | No | ADMET Number: 19 Contents: B, A, D, M, T | - | Yes/No | No |
| vNN-ADMET | Registration required | No | No | ADMET Number: 14 Contents: A, D, M, T | ECFP4 fingerprints | No/ No | No |
| simCyp | free | - | Yes | ADME | PBPK model | - | - |
| SMARTCyp | free or commercial | No | No | Cytochrome P450 | SMARTCyp algorithm | No/No | No |
| MetaPred | free | No | No | Cytochrome P450 and its Isoform | Genetic algorithm SVM (For descriptor selection) | No/No | No |
| P-gp Rules | free | No | No | P-gp Substrate, Pgp inhibitor | Classification and regression tree (CART) algorithm | No/No | No |
Network Pharmacology [15]
Network pharmacology, which integrates information science and systematic medicine, is evolving as a frontier research field of pharmacodynamics HDIs i.e., mode of action. Traditional medicine makes a feature of personalized, holistic and multicomponent therapy, and still plays a key role in modern health care. The systems thinking of traditional medicine have shared much with the core ideas of network pharmacology, which provided important insights for modern drug discovery and may serve as a foundation of future rational drug development against complex diseases. Compared with modern drugs with explicit mechanisms, the challenge for traditional medicine is to understand the molecular mechanism of multicomponent therapies. Moreover, the lack of scientific knowledge of the pharmacological action and the bioactive principle within the multicomponent therapies has already hindered the advance of traditional medicine. Network pharmacology has provided key reference for evidence-based efficacy standards and safety evaluation of traditional medicine. With a network-based insight, network pharmacology intends to systematically reflect and reveal the biological foundation of complex diseases and drug effects. At the same time, “network target”, a key concept that derived from the multi-targets nature of traditional medicine has been proposed, shifting the current “single target” research paradigm. The network-target-based network pharmacology is a promising strategy for the next generation mode of drug research and development for traditional medicine.
Pharmacokinetic HDIs:
Pharmacokinetics is what the body does to the drug/xenobiotic during its fate in the body via four steps namely, absorption, distribution, metabolism and excretion (ADME). Pharmacokinetic HDI can occur at any of these stages via functional alteration of the DMEs and DTs by investigational drugs. These can be evaluated by metabolism-based drug interactions and transporter mediated drug interactions. Metabolism based drug interactions help to identify the enzymes that metabolize an investigational drug and determine if the drug is an enzyme inhibitor or inducer [4]. Various test systems used include but not limited to, subcellular fractions of human liver tissues (microsomes), whole-cell models of isolated human hepatocytes, liver slices, or human cell lines [16]. Transporter mediated reactions tell if an investigational drug is a substrate or an inhibitor of various transporters. Transport activity is generally determined using cell lines such as Caco-2 or MDCK cells that over express specific human transporters, in which bidirectional transport can be measured [17].
Pharmacodynamic HDIs:
Pharmacodynamics is what a drug does to the body. Two concomitantly administered drugs can exert additive, synergistic or antagonistic effects. Accordingly, their dose and duration might change. Pharmacodynamic HDIs can be quantified based on Chou-Talalay or combination index (CI) theorem. According to this, combinations having CI < 1=synergism, CI> 1 = antagonism and CI = 1 = additive effect [18] . CompuSyn (https://www.combosyn.com/) is widely used software for calculating CI.
Pharmacokinetics HDIs:
In vivo studies in humans have been carried out with various experimental designs. Typically, subjects receive a single dose of a test drug or a cocktail of drugs that are markers for various enzymes on day 1. This is followed by multiple daily dose treatment with the herbal product (typically one week) and on the last day of treatment, administration of the test drug or the cocktail of drugs. A comparison of the various pharmacokinetic parameters or phenotypic measures is used as a method to evaluate the effect of herbal products on the pharmacokinetics of test drugs or activity of various drug metabolizing enzymes [19].
Pharmacodynamic HDIs:
This can be a complex study which might depend on the receptor target of the investigational drug, disease condition, drug and route of administration etc.
References:
1. Obodozie OO. Pharmacokinetics and Drug Interactions of Herbal Medicines : A Missing Critical Step in the Phytomedicine / Drug Development Process. Readings Adv Pharmacokinet - Theory, Methods Appl. 2012;127–56.
2. Narayana DA and CK. Draft amendment to drugs and cosmetics rules to license science based botanicals, phytopharmaceuticals as drugs in India. J Ayurveda Integr Med. 2013;4(4):245.
3. Bhatt A. Phytopharmaceuticals: A new drug class regulated in India. pectives Clin Res. 2016;7(2):59.
4. Borse S, Singh D, Nivsarkar M. Understanding the relevance of herb–drug interaction studies with special focus on interplays: a prerequisite for integrative medicine. Porto Biomed J. 2019;4(2):e15.
5. Sharma, A.K., V.K. Kapoor and GK. Herb–drug interactions: a mechanistic approach. Drug Chem Toxicol. 2020;1–10.
6. HU Z. Herb-drug interactions. Drugs. 2005;65(9):1239–82.
7. Mallet L, Spinewine A HA. The challenge of managing drug interactions in elderly people. Lancet (London, England). 2007;370:185–91.
8. SB. BDaJ. Biopharmaceutics and Pharmacokinetics—A Treatise. New Delhi: Vallabh Prakashan; 2009. 140–150 p.
9. Tucker GT, Houston JB HS. Optimizing drug development: Strategies to assess drug metabolism/transporter interaction potentialtoward a consensus. Clin Pharmacol Ther. 2001;70:103–14.
10. Harle U GN. Emerging challenge of herb-drug interaction. Indian J Pharm Educ. 2005;39:71–81.
11. Schneider G. Prediction of drug-like properties InMadame Curie Bioscience Database. Landes Bioscience. 2013. Available from: https://www.ncbi.nlm.nih.gov/books/NBK5974/
12. Hughes JP, Rees SS, Kalindjian SB, Philpott KL. Principles of early drug discovery. Br J Pharmacol. 2011;162(6):1239–49.
13. Sliwoski G, Kothiwale S, Meiler J, Lowe EW. Computational methods in drug discovery. Pharmacol Rev. 2014;66(1):334–95.
14. Pires DEV, Blundell TL, Ascher DB. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–72.
15. Frontiers. Research topic: Network Pharmacology and Traditional Medicine. 2014. Available from: https://www.frontiersin.org/research-topics/7455/network-pharmacology-and-traditional-medicine
16. MacGregor J. In vitro human tissue models in risk assessment: report of a consensus-building workshop. Toxicol Sci. 2001;59(1):17–36.
17. Brantley S. Herb–drug interactions: challenges and opportunities for improved predictions. Drug Metab Dispos. 2014;42(3):301–17.
18. Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22(C):27–55.
19. Venkataramanan R, Komoroski B, Strom S. In vitro and in vivo assessment of herb drug interactions. Life Sci. 2006;78(18):2105–15.
Copyright © Savitribai Phule Pune University, School of Health Sciences, Pune. All Rights Reserved.
[Best viewed in IE 10+, Firefox, Chrome, Safari, Opera.]
:::| powered by dimakh consultants |:::